

It is as if we sequenced the genome and forgot everything else happening within the cell.
 01
01It was a watershed moment when we sequenced the first human genome in April 2003. This milestone was quickly followed by Next-Generation Sequencing (NGS) in 2006 and RNA-Seq in 2008. Almost two decades later, our ability to map the genetic code has transformed the life sciences. It has given us paradigm-shifting precision medicine, world-saving mRNA vaccines, fortune-rewriting gene therapy, Nobel-winning gene editing, and cancer-busting cell therapy. I recently attended a conference where Bill Kaelin, the 2019 Nobel Prize winner, opined that we have barely scratched the surface of potential discoveries based on life’s genetic template. I couldn’t agree more.
As a scientist, imagining a world without these genome-centric tools is difficult. At the same time, viewing the life sciences nearly exclusively through the lens of the genome is one of our most significant limitations. As Patrick Malone put it recently: “In science, we frequently confuse the map for the terrain.” To extend Patrick’s analogy, we used sequencing “cartography” to build a “map” of biology based on the genome and its products (RNA and proteins) and nonchalantly operate as if this map accurately and faithfully recapitulates the entire terrain of biology. It is as if we sequenced the genome and forgot everything else happening within the cell. What maps of life could be built outside of those enabled by the genetic template? What maps should we build?
 02
02A great way to answer the questions above is to ask what comes after genes, RNA, and proteins. Cells are considered the fundamental units of life, and proteins are their primary workhorses. But what “work” do these proteins do for the cell? Borrowing the words of legendary biophysicist Harold Morowitz, they make energy flow and matter cycle.
Let’s take the infamous cancer protein target, KRAS, for example. KRAS is a poster child for a genetic worldview of health and disease. It is the fearsome “Death Star”-shaped general to the emperor of maladies, cancer. What does KRAS actually do? Mutant KRAS, it turns out, alters glucose metabolism to promote glucose uptake by cells and induces the expression of specific kinases that are rate-limiting enzymes of glycolysis. It also promotes a special biosynthesis pathway called the hexosamine biosynthesis pathway, which provides precursors for lipid and protein glycosylation. It simultaneously activates the bespoke non-oxidative arm of the pentose phosphate pathway, which supplies ribose, the backbone for nucleic acid production (cancer’s growth, unsurprisingly, involves doubling of the cell’s contents; it involves producing and assembling lipids to create more cell membranes, amino acids to manufacture more proteins, and nucleic acids to form yet more affected genetic material). There’s more! KRAS mutants rewire glutamine metabolism, a critical metabolite that along with glucose, is sufficient for cancer growth in cell culture experiments. While at it, and for good measure, KRAS also stimulates scavenging pathways such as macropinocytosis and autophagy, providing yet more building blocks to the anabolic synthesis routes. It does all this while subtly maintaining appropriate energy levels and the cell redox potential so as to not set off deeply ingrained cellular anti-cancer defenses. Like all proteins, KRAS choreographs the intimate dance of chemical bond-breaking and bond-making to sculpt the pathways through which our cells shuttle carbon and energy.
As Nick Lane, one of the pre-eminent voices of the now eclipsed world of biochemistry, writes in his recent book, Transformer: The Deep Chemistry of Life and Death, “Information isn’t life…a dead cell and a live cell have the same genome”. Instead, he contends, that metabolic flux —- the sum of continuous transformations of small molecules on a timescale of nanoseconds, nanosecond after nanosecond —- is the definition of being alive. Life is the ability of cells to regenerate themselves continually from simpler building blocks. I find it hard to disagree with Nick.
 03
03Chemistry links all life more intimately than genetics. The glorious thesis that cells are animated by the flow of energy and the cycling of matter is valid in all cells. How bacteria respire with oxygen or grow in its absence is remarkably similar to how our heart cells behave in different environments. In the 1920s, Albert Kluyver, the great Dutch microbiologist, exclaimed, “From Elephants to Butyric acid bacterium – it’s all the same!” to marshall the thinking that fundamental biochemical pathways such as the Krebs Cycle are conserved across all cells. This observation was the first universal code of life accepted by science long before its more popular counterpart – the one that spawns the infinite symphonies of twenty amino acids across life forms.
If we reframe life as metabolism, then our map of life is woefully incomplete. Today, even with our most sophisticated tools to annotate (i.e., identify or name) the masses obtained from an untargeted metabolomics experiment, we can barely annotate 6% of the metabolites in human plasma (see Figure 1 below). To borrow a comparison from our scientific co-founder, Pieter Dorrestein, our ability to annotate the fraction of known genes in bacteriophage metagenomes (a well-known frontier of uncharacterized dark biological matter) can be as high as 30%. In other words, we know more about the genetics of an uncultured virus than we do about the chemistry of our bodies. Suppose we are barely scratching the surface of genetics-driven discovery. In that case, we can scarcely even see the surface of chemistry.
Figure 1: Representative metabolites found in human plasma arranged by a similarity fingerprint. Highlighted in red are molecules known to science. Data generated in Enveda’s labs.
Even beyond human blood, about 5% of the molecules within any natural sample (think plants, microbes, fungi, etc.) are currently identifiable. Moreover, the annotated ~5% is largely shared between many organisms, putting optimistic estimates of the total annotated chemistry of our world at <<1%. The total of metabolites known to humankind is a mere ~400,000. This number includes all the chemistry in us, on us, and around us.
Nothing illustrates the blue-sky opportunity of chemistry-driven discovery more than a series of groundbreaking discoveries in Pieter’s lab. Focused on a tiny portion of the signal from a metabolomics experiment of mammalian gut samples, Pieter’s lab discovered puzzling molecules resembling both bile acids and amino acids. They had found three new bile-acid conjugates unknown to science. This discovery came nearly a quiet century after Heinrich Wieland’s Nobel prize in 1927 for what was then assumed to be a “comprehensive” description of bile acid chemistry. Since that Nature publication in 2020, Pieter’s team has already described hundreds of unknown bile acid structures in various organs (see Figure 2). Unsurprisingly, they observed stark changes in these bile acid profiles across many diseases. Zooming out, they might be onto something even more significant. Given the unique pattern of these bile acid structures, their modifying enzymes, and receptors within various organs, they posit that bile acid conjugation is the code that ensures high-specificity delivery of particular metabolite cargo to target organs. They may have just solved a fundamental question in biology: how is matter cycled efficiently in an organism, given the varying demands of different specialized organs?
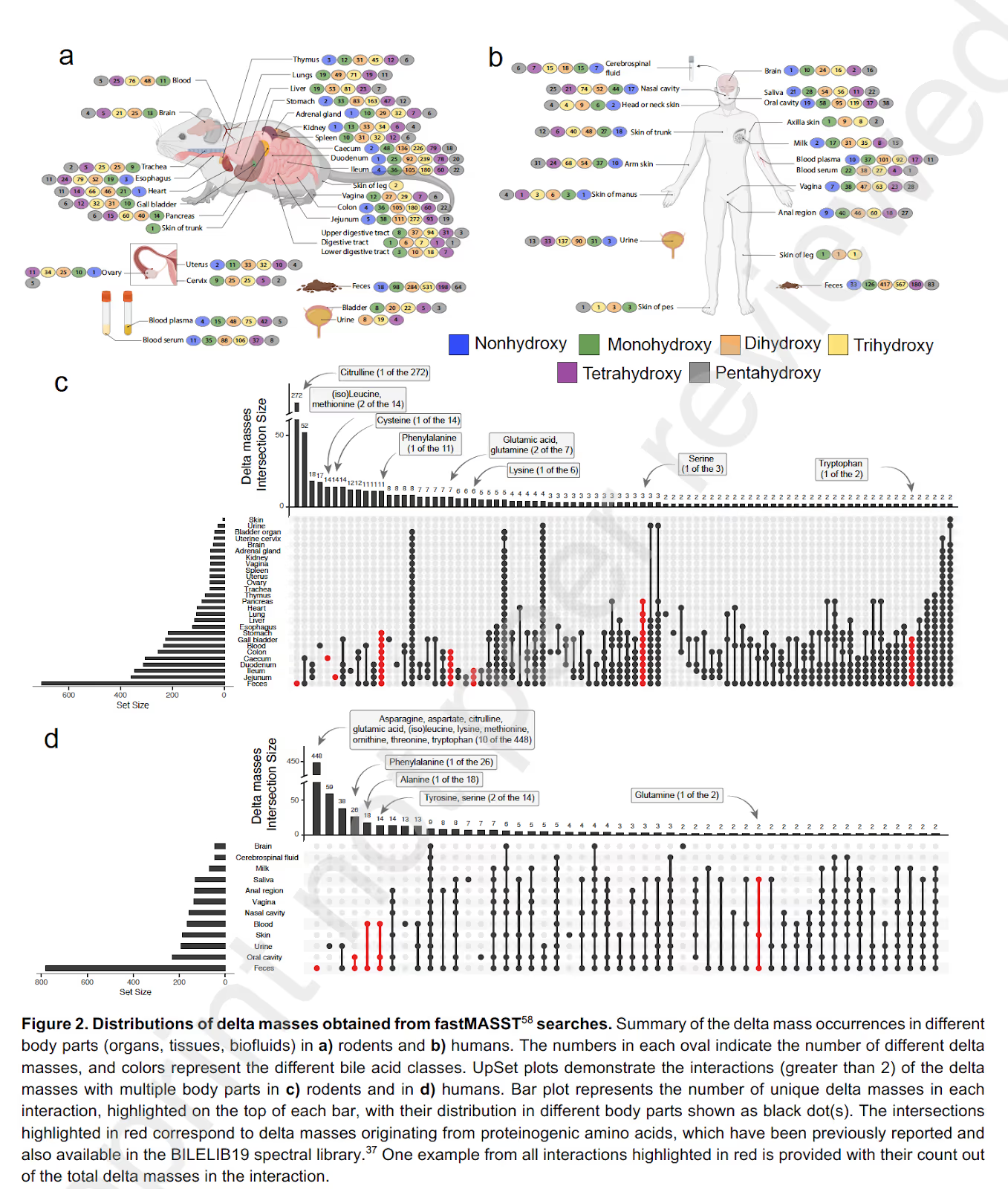
Figure 2: Showing various bile acid conjugates in rodent and human samples in analyses performed by the Dorrestein lab. Figure adapted from this preprint.
 04
04While the work from the Dorrestein lab is exciting, it was also hard-fought, requiring targeted effort to untangle and identify this class of novel metabolites. To comprehensively crack the chemical code of life, we need an “NGS moment” for metabolomics. Today, we can take any sample with genetic material and (i) determine the sequence of nucleic acids to ask what the genes are, and (ii) compare them to other sequences to ask what the genes might do. The reliance on this genetic template is critical for our ability to analyze our transcriptomes and proteomes.
We lack such a “sequencer” technology for the world’s chemistry. When we founded Enveda in 2019, the only way to characterize unknown units of the world’s chemistry was one molecule at a time. One needed to purify individual molecules, obtain sub-milligram quantities of each and perform lengthy experiments using a technique called NMR Spectroscopy to determine the structure of each molecule. Once that analysis, which can take weeks or months, was complete, the pure molecule could be retrieved and tested in individual biological experiments to determine its putative function. This process is akin to sequencing genetic code one base pair at a time.
At Enveda, we knew we had to solve this grand challenge to unlock the latent potential of our planet’s chemistry. We had two choices to answer what the molecules were and what they did. Invent a groundbreaking analytical method akin to NGS or re-examine existing analytical technologies that allow us to generate representations of the chemistry contained within any sample. We chose the latter, which led us directly to tandem mass spectrometry (LC-MS/MS). LC-MS/MS allows us to generate rich representations of individual molecules after ingesting highly complex mixtures. It generates these “spectra” (see Figure 4) by using an electric field to accelerate the ions and slam them into a neutral gas so that they wiggle until the weakest bonds start to break. It then captures the masses of the resulting fragments to generate a “fingerprint.” The problem was that these spectra were used simply as fingerprints – scientists would look for spectral fingerprint matches to unearth known chemistry in their sample. As you might imagine, the problem with this approach is that one can only rediscover known chemistry. As I mentioned earlier, this known chemistry is just a minuscule fraction of the whole.
We wanted to challenge that use. A little over two years ago, we found that these fingerprints were a perfect match for a specific kind of machine-learning methodology called Transformers. It turns out nature’s chemistry has a grammar, and we can use it to translate spectra into a language humans can understand: chemical structure and molecular properties. To learn more about how we built these models, see here. Today, we are already nearly three times better at predicting the properties of >95% unknown molecules in a sample (see Figure 3 below and full paper here) and twice as good at predicting approximate structure, both compared to looking up the closest fingerprint match (see here). Importantly, we can do this at thousands of predictions per second and are improving weekly. In parallel, we have built technologies that allow us to identify active and inactive molecules in the context of any assay in one single step, reducing years of overall effort into several hours. With these tools applied at large, we are on our way to building the most comprehensive map of the world’s chemistry.
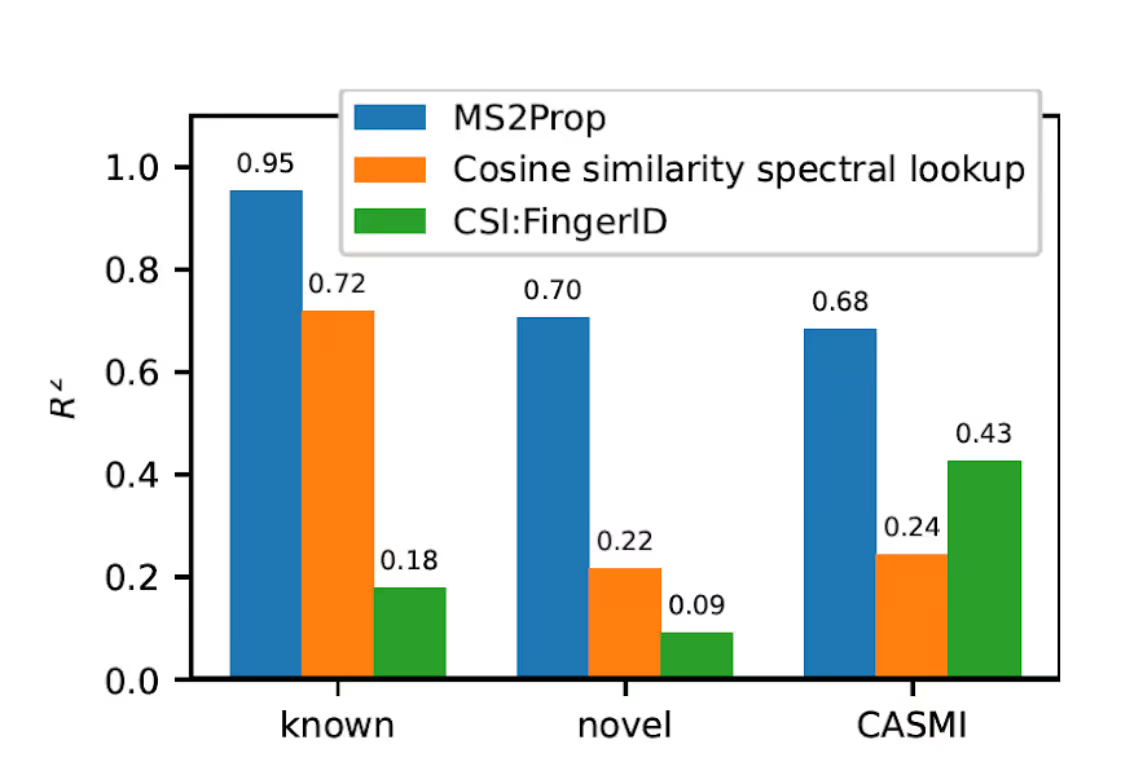
Figure 3: MS2Prop performance as measured by R2 compared to looking up the closest fingerprint (orange) and a computational tool to predict structures from this paper.
Figure 4: MS2Mol translates spectra into chemical structures, much like ChatGPT is able to translate between two languages.
 05
05What would we do with such a map? What could we do with the most advanced sequencer for the world’s chemistry? The possibilities, in my view, are limitless. With the recurring theme invoked by the pioneering Leslie Orgel, “Evolution is cleverer than you are,” let’s explore a few big ideas within our reach at Enveda.
Unlocking the world’s most powerful library for modern drug discovery: As Joe Rokicki, our brilliant founding CTO, once said, “Every metabolite in nature is made by a protein and for a protein.” Considering shared evolutionary pressures that lead to repeating patterns of protein shape (a study of 20,000 ligand binding pockets showed that about 1000 shapes could represent the structural space), one would expect protein-made metabolites to be bioactivity enriched. Comprehensively sampling metabolite chemistry is perhaps the most efficient way to cover the vast universe of possible small-molecule chemical space (posited as high as 10^60 compounds) relevant for biological activity. Just about two dozen discovery campaigns in, we already see significant signs of this evolutionary relevance at work at Enveda. We have had remarkable success finding ligands for “undruggable proteins,” modulators of protein-protein interactions, and effectors of completely new biology derived from phenotypic screens unexplainable with known targets. It is worth mentioning that the moniker undruggable is derived from unsuccessful screening efforts involving synthetic libraries. It is unsurprising, then, that a vastly more relevant and diverse library would be able to deliver hits against those targets. Another essential aspect would be the utility of this evolutionary relevance in providing a therapeutic effect in the form of medicine. The property space of metabolites is associated with lower failure rates in preclinical toxicology studies. Moreover, our analyses have shown that metabolite libraries cover ~3X more FDA-approved drug space with 1/10th of the compounds than synthetic libraries (see Figure 5). In the words of our phenomenal Head of Platform Chemistry, Marvin Yu, not all metabolites are drug-like, but all drugs are metabolite-like.
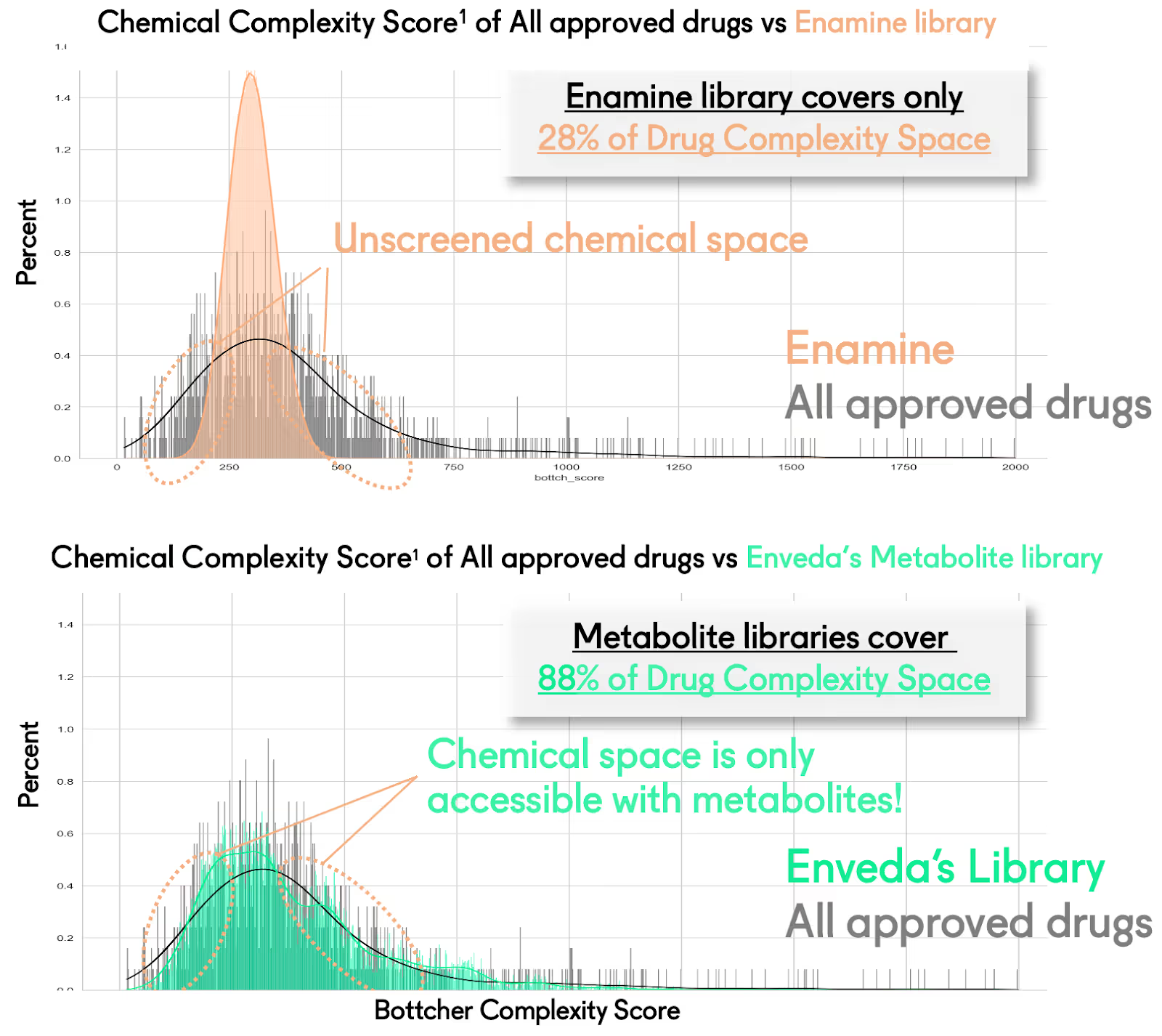
Figure 5: Comparison of chemical space (as defined by Bottcher complexity score) covered by 2M compounds from Enamine REAL versus 200,000 metabolites in Enveda’s library.
Understanding the chemical basis of disease: As sequencing-based omics technologies were invented, scientists quickly applied them to create disease signatures of genes, transcripts, and proteins. Analyses of these signatures have led to the discovery of novel disease targets, pathological mechanisms, and the subsequent identification of drugs that reverse these signatures. However, this work relies on biological representations in the form of genes or their products. At Enveda, we can’t help but wonder what a chemical representation of our most challenging diseases would look like. As cancer cells acquire resistance mutations, do they maintain their metabolic rewiring? If so, will a therapeutic that reverses the metabolic dysfunction provide a more promising avenue to treat cancer without inducing rapid resistance? What would medicines discovered using metabolomic fingerprints look like? In a breakthrough study published in Nature recently, a team from Stanford identified a novel metabolite induced by exercise (in humans and racehorses!) that could help fight obesity when administered to overweight mice.
Detecting, stratifying, and monitoring disease using metabolites:Since chemistry is where the action happens, it should also faithfully capture any deviation from the steady state. Such variation is especially key when pathological mechanisms are clearly downstream of the genome. Identifying the mutations in a tumor’s DNA has been game-changing for cancer treatment, but this approach is conceptually limited to cancer. For example, the genetic code of the inflamed gut in a patient suffering from inflammatory bowel disease (IBD) is the exact same as the rest of the body. What could the metabolites in their guts tell us about their disease? Numerous academic studies have reported disease-induced metabolome changes across IBD and various other diseases, including those that have traditionally been difficult or cumbersome to diagnose (see here, here, here, and here for examples). We are excited to extend these academic observations into robust solutions that identify and stratify patients based on their chemical signatures. Coupled with therapeutic candidates that reverse disease signatures in the laboratory (effort #2 above), we imagine an era of precision medicine fueled by our chemistry rather than our genes.
 06
06The benefit that science, medicine, and society have accrued from the human genome sequence and advancing sequencing technologies is undeniable. But to believe that this is the entire terrain of cell and organismal biology severely limits our ability to understand life. By creating technologies that unlock nature’s chemistry, like sequencing unlocked genomics, we are creating a new and complementary map of biology. With the chemical code in our grasp, Enveda will usher in a new era of human and planetary health.